
Galaxy is an open, web-based platform for data-intensive computational research that spans beyond the life sciences. It allows researchers without programming experience to run analysis workflows on their data, share their results, and enable others to repeat the same analyses. Galaxy makes science reproducible, facilitates sharing of data and results, and removes the hassle of installing software tools from users.
ELIXIR's Galaxy Community evolved from its Galaxy Working Group, which was established in 2015 to monitor and foster the use of Galaxy in ELIXIR. Today the overall goal of the Community is to foster the use and development of Galaxy, focusing on making it easier to import data into Galaxy instances, helping to develop and share Galaxy tools and workflows, and increasing the provision of Galaxy training.
Goals of the Community
To grow the European network of Galaxy communities
- An increasing number of scientific sub-communities have grown up to address specific tasks using Galaxy. The collaborative portal Workflow4Metabolomics (W4M) and GalaxyP, for example, are dedicated to handling metabolomic data and feature in the work of the ELIXIR Metabolomics Community.
- The Galaxy Community's goal is to foster interactions between the domain-specific Galaxy communities, to set up common analysis workflows and standards, and to provide training in these.
- The Galaxy Community acts as an umbrella organisation for many regional communities in France, UK, Netherlands, Czech Republic, etc., and supports other groups in forming such groups.
To grow the global network of Galaxy communities through partnership, contribution and leadership
- The Galaxy network is a globe-spanning, highly impactful digital research infrastructure, used by hundreds of thousands of researchers across all continents. This infrastructure is built and sustainably supported through the efforts of hundreds of contributors and resource providers. Mutual partnership and agreement between global Galaxy providers are necessary for the open, rapid and continuous development of Galaxy functionalities, tools, workflows and all the other content, which in turn benefit all participating providers and communities.
- Therefore an important goal of the ELIXIR Galaxy Community is the growth of the global Galaxy community, through leadership, governance, content development, platform sustainability, shared resources, community fostering, and training.
- This goal can be realised through close coordination and partnership with existing Galaxy provider peers including the US, Australia and South Africa; and then through broader networking and leadership globally.
- Galaxy plays a crucial role in different EOSC consortia (EOSC-Life, EOSC-Nordic, EOSC-Pillar), supporting a wide variety of use cases across different disciplines; and will continue participating in different European initiatives.
- The usegalaxy.* servers across Europe and the world are offering access to 2900+ tools, workflows and national, as well as international compute resources.
To extend Galaxy training provision
- ELIXIR-organised workshops are having a fundamental impact in raising the profile of the Galaxy Training Network (GTN) among global bioinformatics training efforts. This is a training material repository open for everyone to use and contribute to, providing slides, tutorials and other material on using, developing and administering Galaxy. We aim to keep these materials up-to-date and expand them to cover further areas of the life sciences. See the GTN Statistics for an overview of the topics addressed and other analytics.
- usegalaxy.* will offer Training Infrastructure as a Service (TIaaS) – read some feedback about TIaaS on usegalaxy.eu from the instructors.
- We will continue promoting the use of these resources, including information about where these trainings can be run, trainers, needed tools and virtual Galaxy images. This will be done in collaboration with the Training, Tools and Compute Platforms.
To create a Galaxy cloud infrastructure across Europe

- The growing amount of data generated in life science, and the large number of Galaxy communities, requires increasing compute and storage resources. Our aim is to facilitate access to a broad portfolio of analysis workflows for European researchers.
- Some ELIXIR Nodes already offer a centralised instance of Galaxy (France, Germany, Belgium, Norway, Spain). Since 2018, in collaboration with the US Galaxy Team, we are building a network of Galaxy instances worldwide (usegalaxy.*), guaranteeing a base level of compatibility and supporting all training materials of the Galaxy Training Network. See the flyer for usegalaxy.eu for more information.
- We also want to facilitate the usage of Galaxy on top of the different ELIXIR clouds, e.g. by using CloudLaunch as a single entry point for users; or through technologies and services supporting the deployment of Galaxy on federated cloud infrastructures such as Laniakea. ELIXIR-Italy launched the service Laniakea@ReCaS, hosted at the ReCaS-Bari datacenter, providing cloud resources for the deployment of on-demand Galaxy instances.
To make it easier to access and transfer data
- Getting data from public databases into a Galaxy instance is the first step for most analyses. However, identifying files and their URLs and uploading these files in a computational environment is not easy for users with limited technical skills.
- We aim to facilitate uploading data into Galaxy instances from the ELIXIR Core Data Resources such as ENA, EGA, ArrayExpress, PRIDE and UniProt, and also from more specialised databases such as Brenda, Silva and RNACentral. To optimise data access and integration in Galaxy we need to standardise and automate data transfer.
- We are working closely with the worldwide Galaxy community to create and maintain shared storage of common reference data for numerous genomes, to be used across Galaxy instances via the CVMFS technology. This facilitates the inclusion of new reference genomes to any Galaxy instance, immediately providing indices and annotations.
To improve tools and data integration
- Currently, a data-to-tools approach is prevalent in data analysis. This involves copying a large volume of data to a computing environment for analysis. To avoid this, we propose a tools-to-data approach based on virtualization, such as Docker, Singularity or rkt.
- Galaxy already supports BioContainers, meaning that tools and workflows in Galaxy can run in isolated BioContainers. We aim to maintain, update and extend BioContainers integration to keep the resource relevant and up-to-date.
- We also aim to improve the accessibility of tools and data, allowing users to easily combine public and private storage and compute cloud services.
To promote FAIR principles in Galaxy
- The ELIXIR Galaxy Community will promote the use of Galaxy projects that enhance the FAIRness of Galaxy. These include Galaxy ToolShed (a repository of Galaxy tools and utilities) and GalaxyCat (an online catalogue of the tools available on various Galaxy instances).
- We will promote the use of the ELIXIR Tools registry bio.tools and work with bio.tools developers to integrate BioConda, BioContainers and the Galaxy ToolShed more tightly into the registry.
- The Galaxy Community aims to work with the Interoperability Platform to annotate Galaxy objects (histories, workflows, etc.) as standardised ResearchObjects to facilitate sharing.
Accomplishments
Compute infrastructure
- The Pulsar Network allows the distribution of jobs across different data centres in Europe.
- More information about the user statistics can be found on the Grafana page of the European Galaxy Server and the factsheet.
Integration with ELIXIR resources
- We enabled easy authentication on Galaxy instances by using the ELIXIR AAI. This was achieved in close collaboration with the Compute Platform in ELIXIR.
- Galaxy workflows available in the WorkflowHub can be run now directly on usegalaxy.eu.
Training
- Galaxy training material is integrated and listed in TeSS, and is annotated with BioSchemas markup.
- Online training has become more prominent since the COVID-19 pandemic started. The Galaxy community has organised several massive online trainings and our experience and recommendations are gathered in this paper.
- Webinars on Galaxy Advanced Features.
- Training events are happening all the time, check the upcoming ones.
Response to the COVID-19 pandemic
- Two webinar series have taken place since the spring of 2020:
- From early 2020 the Galaxy Community has collaborated across the globe to provide tools, workflows and access to data related to COVID. All the news related to the latest COVID-19 data analysis is available in a collection of blog posts. The major achievements are:
- Genomics, Proteomics, Evolution, Cheminformatics and Protein-Protein Interaction analysis can be found at the Galaxy COVID-19 website.
- Submission of viral data to the COVID-19 Data Portal through Galaxy.
- Collaboration with Viral Beacon to visualise and offer results from variant analysis workflows.
- In 2021, an automated monitoring of new data was set up in response to the evolving pandemic.
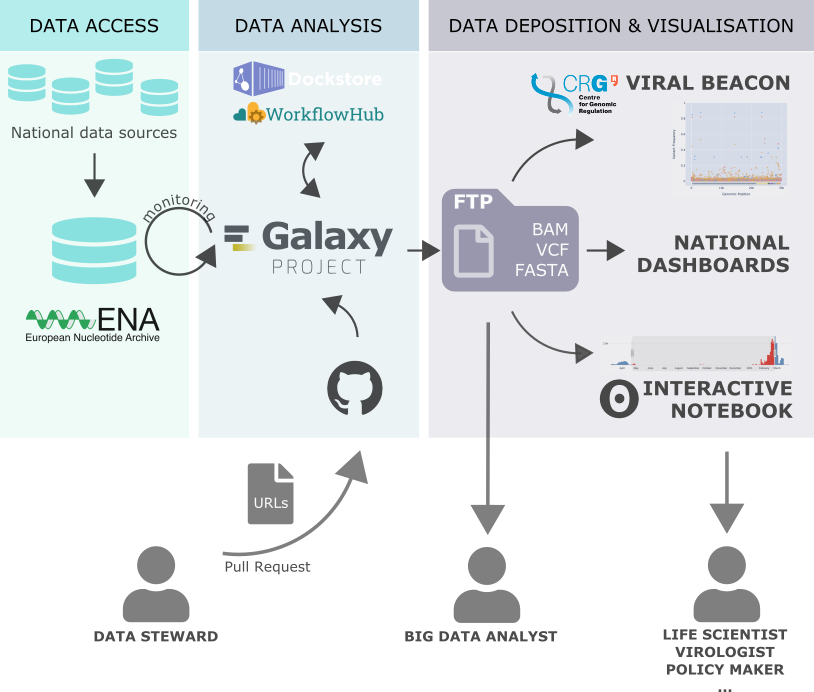
- All the activities are summarised and periodically updated in this running document.
- Publications
- Baker D, van den Beek M, Blankenberg D, Bouvier D, Chilton J, et al. (2020) No more business as usual: Agile and effective responses to emerging pathogen threats require open data and open analytics. PLOS Pathogens 16(8): e1008643. https://doi.org/10.1371/journal.ppat.1008643
- Gallardo-Alba C, Grüning B, Serrano-Solano B (2021) A constructivist-based proposal for bioinformatics teaching practices during lockdown. PLOS Computational Biology 17(5): e1008922. https://doi.org/10.1371/journal.pcbi.1008922
- Serrano-Solano B, Föll MC, Gallardo-Alba C, Erxleben A, Rasche H, et al. (2021) Fostering accessible online education using Galaxy as an e-learning platform. PLOS Computational Biology 17(5): e1008923. https://doi.org/10.1371/journal.pcbi.1008923
- Rajczewski,A.T. et al. (2021) A rigorous evaluation of optimal peptide targets for MS-based clinical diagnostics of Coronavirus Disease 2019 (COVID-19). Clinical Proteomics, 18, 15. https://doi.org/10.1186/s12014-021-09321-1
Community
- Check the latest news and events. There are community-focussed events happening all year long, like Developer Roundtables, CoFests and the annual Galaxy Community Conference (GCC).
- The Galaxy community is very active and spread all over the world. Check how to become part of it in the following video:
Commissioned Services
The Galaxy Community has been involved in a number of short-term, technical projects called Commissioned Services. For a complete list of finished and ongoing Implementation Studies, see the Commissioned Services page.
Leadership

(ELIXIR UK)
Community co-lead

(ELIXIR Germany)
Community co-lead

(ELIXIR UK)
Community co-lead

Mihail Anton
(ELIXIR Hub)
Community Liaison
Find out more
- For Galaxy activities from 2015-18 see the Galaxy Working Group page.
- Publications:
- Doppelt-Azeroual, O., Mareuil, F., Deveaud, Kalaš, M., Soranzo, N., van den Beek, M., Grüning, B., Ison, J. and Ménager, H. (2017). ReGaTE: Registration of Galaxy Tools in Elixir GigaScience, doi:10.1093/gigascience/gix022
- Contact galaxy-coleads@elixir-europe.org or galaxy-wg@elixir-europe.org if you'd like to know more about the Community's work.